-Search query
-Search result
Showing 1 - 50 of 134 items for (author: finley & m)

EMDB-16964: 
MutSbeta bound to (CAG)2 DNA (canonical form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

PDB-8olx: 
MutSbeta bound to (CAG)2 DNA (canonical form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-16972: 
MutSbeta bound to 61bp homoduplex DNA
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

PDB-8oma: 
MutSbeta bound to 61bp homoduplex DNA
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-16971: 
MutSbeta bound to (CAG)2 DNA (open form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

PDB-8om9: 
MutSbeta bound to (CAG)2 DNA (open form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-16973: 
DNA-unbound MutSbeta-ATP complex (bent clamp form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

PDB-8omo: 
DNA-unbound MutSbeta-ATP complex (bent clamp form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-16974: 
DNA-unbound MutSbeta-ATP complex (straight clamp form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

PDB-8omq: 
DNA-unbound MutSbeta-ATP complex (straight clamp form)
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-16975: 
MutSbeta bound to homoduplex plasmid DNA
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-16969: 
DNA-free open form of MutSbeta
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

PDB-8om5: 
DNA-free open form of MutSbeta
Method: single particle / : Lee JH, Thomsen M, Daub H, Steinbacher S, Sztyler A, Thieulin-Pardo G, Neudegger T, Plotnikov N, Iyer RR, Wilkinson H, Monteagudo E, Felsenfeld DP, Haque T, Finley M, Dominguez C, Vogt TF, Prasad BC

EMDB-14083: 
26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS)
Method: single particle / : Hung KYS, Klumpe S, Eisele MR, Elsasser S, Geng TT, Cheng C, Joshi T, Rudack T, Sakata E, Finley D

PDB-7qo4: 
26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS)
Method: single particle / : Hung KYS, Klumpe S, Eisele MR, Elsasser S, Geng TT, Cheng C, Joshi T, Rudack T, Sakata E, Finley D

EMDB-32281: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SB_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
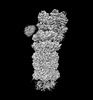
EMDB-32284: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD5_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3i: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SB_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3m: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD5_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
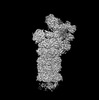
EMDB-32272: 
Structure of USP14-bound human 26S proteasome in state EA1_UBL
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32273: 
Structure of USP14-bound human 26S proteasome in state EA2.0_UBL
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
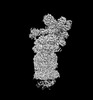
EMDB-32274: 
Structure of USP14-bound human 26S proteasome in state EA2.1_UBL
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
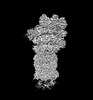
EMDB-32275: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
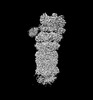
EMDB-32276: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED5_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
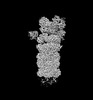
EMDB-32277: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
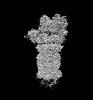
EMDB-32278: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED1_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
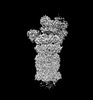
EMDB-32279: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
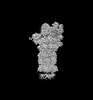
EMDB-32280: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
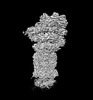
EMDB-32282: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y
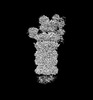
EMDB-32283: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD4_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32285: 
Structure of USP14-bound human 26S proteasome in state EA1_UBL with the local RPN1 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32286: 
Structure of USP14-bound human 26S proteasome in state EA2.0_UBL with the local RPN1 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32287: 
Structure of USP14-bound human 26S proteasome in state EA2.1_UBL with the local RPN1 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32288: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14 with the USP14 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32289: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14 with the RPN1 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32290: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14 with the RPN1 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32291: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14 with the USP14 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-32292: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14 with the USP14 density improved
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w37: 
Structure of USP14-bound human 26S proteasome in state EA1_UBL
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w38: 
Structure of USP14-bound human 26S proteasome in state EA2.0_UBL
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w39: 
Structure of USP14-bound human 26S proteasome in state EA2.1_UBL
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3a: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED4_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3b: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED5_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3c: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED0_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3f: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED1_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3g: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.0_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3h: 
Structure of USP14-bound human 26S proteasome in substrate-engaged state ED2.1_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3j: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SC_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

PDB-7w3k: 
Structure of USP14-bound human 26S proteasome in substrate-inhibited state SD4_USP14
Method: single particle / : Zhang S, Zou S, Yin D, Wu Z, Mao Y

EMDB-14082: 
Structure of the 26S proteasome-Ubp6 complex in the si state (Core Particle and Lid)
Method: single particle / : Hung KYS, Klumpe S, Eisele MR, Elsasser S, Geng TT, Cheng TC, Joshi T, Rudack T, Sakata E, Finley D
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
